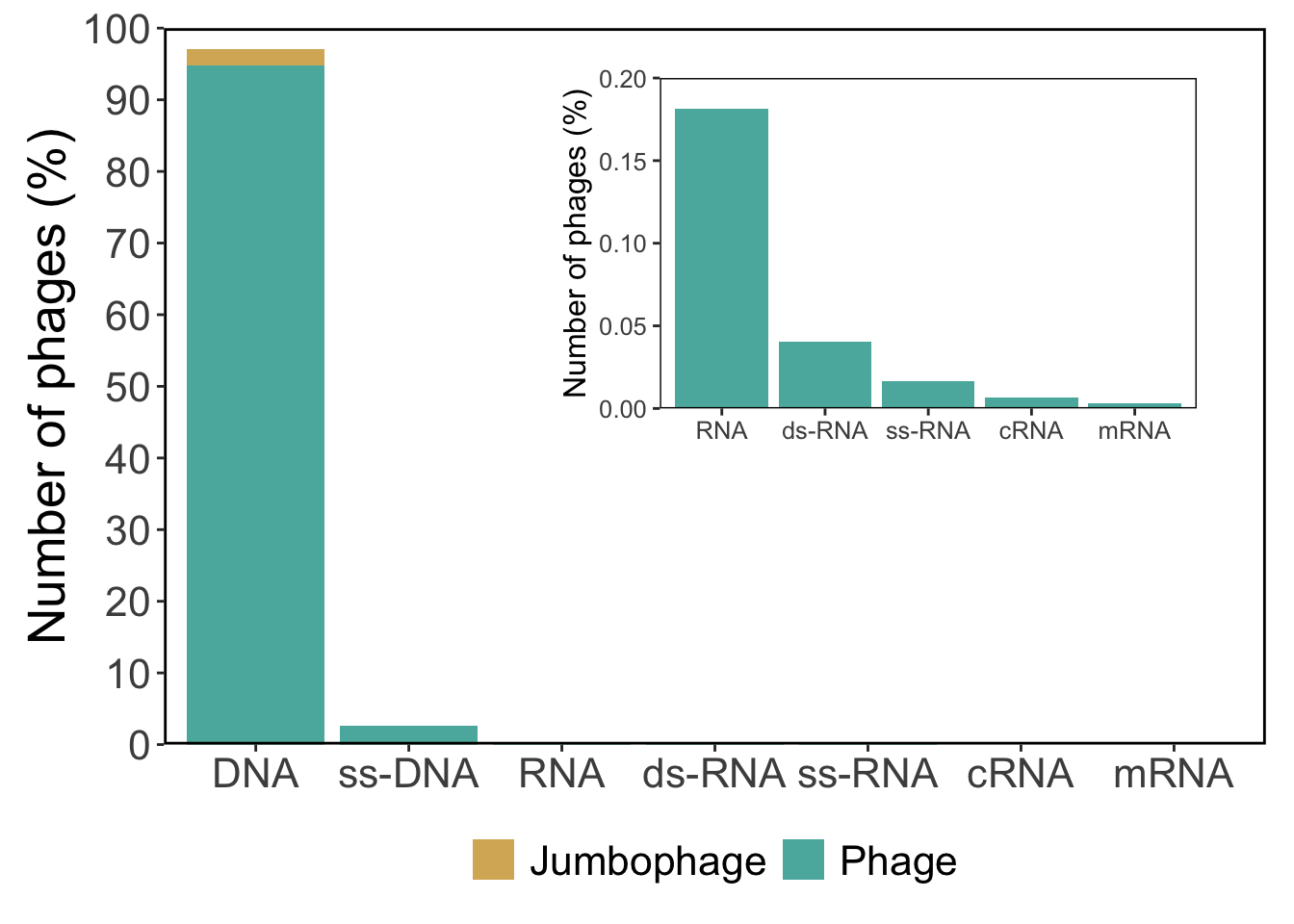
library (tidyverse)library (patchwork)<- read_tsv ("rawdata/clean_genomes_data.tsv" ) %>% select (molecule, jumbophage) %>% count (molecule, jumbophage) %>% mutate (percent = 100 * (n / sum (n)),jumbophage = if_else (jumbophage == FALSE , "Phage" ,"Jumbophage" molecule = factor (molecule,levels = c ("DNA" , "ss-DNA" , "RNA" , "ds-RNA" , "ss-RNA" , "cRNA" , "mRNA" )<- c ("#d8b365" , "#5ab4ac" )<- df %>% ggplot (aes (x = molecule,y = percent,fill = jumbophage+ geom_col () + scale_y_continuous (expand = c (0 , 0 ),limits = c (0 , 100 ),breaks = seq (0 , 100 , 10 )+ scale_fill_manual (values = base_color) + labs (y = "Number of phages (%)" ,x = NULL + theme (text = element_text (size = 20 ),panel.background = element_blank (),panel.border = element_rect (colour = "black" , fill = NA , linewidth = 1 legend.position = "bottom" ,legend.title = element_blank ()<- df %>% filter (! molecule == "DNA" & ! molecule == "ss-DNA" ) %>% ggplot (aes (x = molecule,y = percent,fill = jumbophage+ geom_col () + scale_y_continuous (expand = c (0 , 0 ),limits = c (0 , 0.2 ),breaks = seq (0 , 0.2 , 0.05 )+ scale_fill_manual (values = "#5ab4ac" ) + labs (y = "Number of phages (%)" ,x = NULL + theme (text = element_text (size = 12 ),panel.background = element_blank (),panel.border = element_rect (colour = "black" , fill = NA , linewidth = 0.5 legend.position = "none" + inset_element (p2, left = 0.35 , bottom = 0.4 , right = 0.95 , top = 0.95 )