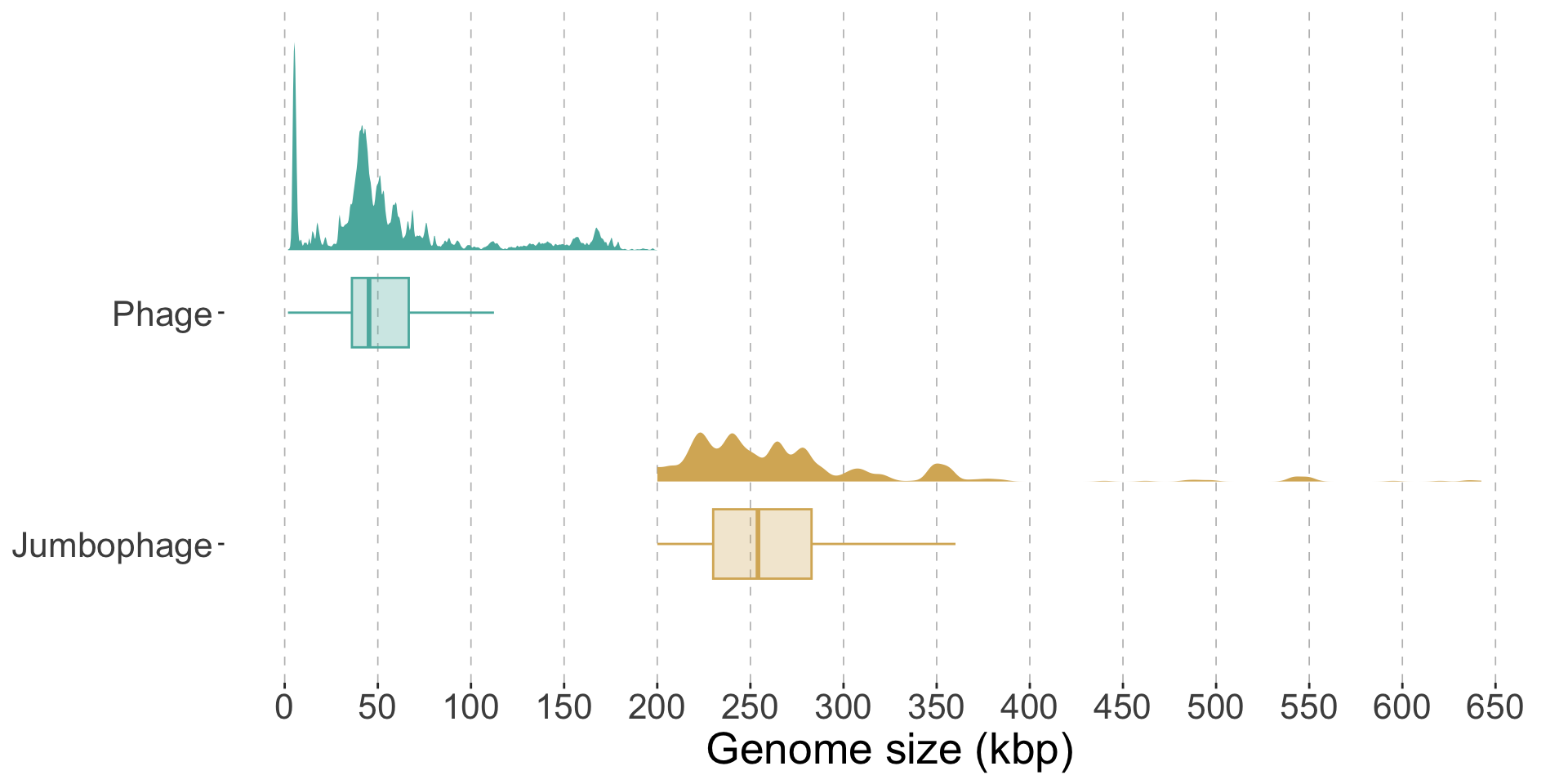
library (tidyverse)library (ggdist)<- c ("#d8b365" , "#5ab4ac" )read_tsv ("rawdata/clean_genomes_data.tsv" ) %>% select (genome_length_bp, jumbophage) %>% mutate (jumbophage = if_else (jumbophage == FALSE ,"Phage" ,"Jumbophage" genome_length_bp = genome_length_bp / 1000 ) %>% ggplot (aes (y = jumbophage,x = genome_length_bp+ :: stat_halfeye (aes (fill = jumbophage),adjust = .5 ,width = 0.1 ,.width = 0 ,justification = - .3 ,point_colour = NA ,color = "black" + geom_boxplot (width = .3 ,outlier.color = NA ,alpha = 0.3 ,aes (color = jumbophage,fill = jumbophage+ scale_x_continuous (limits = c (0 , 650 ),breaks = seq (0 , 650 , 50 )+ scale_colour_manual (values = base_color) + scale_fill_manual (values = base_color) + labs (y = NULL ,x = "Genome size (kbp)" + theme (text = element_text (size = 20 ,color = "black" panel.background = element_blank (),panel.grid.major.x = element_line (color = "grey" ,linewidth = 0.3 ,linetype = 2 # panel.border = element_rect(colour = "black", fill = NA, linewidth = 1), legend.position = "none"